Python
Python is also an intuitive and easy-to-understand programming language that is quickly growing in popularity in the scientific community, due in no small part to how accessible it is to people that do not have a programming background. Python was chosen for several reasons:
- With the growing popularity of python, many process scientists know or can pick up python already
- CC-Sim was written with machine learning in mind; the data output is easily piped to machine-learning frameworks such as Tensorflow and Pytorch
- The experimental setup and simulation can all be automated in python, allowing for automated processes (or even the output of a ML process) to easily define and run a simulation
Modular Controls
Controls make up a huge amount of variance between processes. They are easy to customize here so that the digital process mimics a real process. There’s built-in controls for common things like aeration, heating, and feed, with customization ranging from defining what media to connect to the feed to PID constants. If a control can be described with a set of equations, it can be programmed into python.
Each control takes a set of observed values (from the assays module, to ensure no data leakage) and outputs the signal that would be sent to actuation. From there, actuation determines what physical changes actually occur and affect the environment.
Random Variance
Variation occurs wherever we would expect there to be random environmental input in the process. Each batch of media will vary from its intended definition; each assay may include systematic, drift, and random errors; each actuation component will vary from the intended signal. Variation is even included in sampling times and, optionally, components breaking. All of this adds up to create the variation we see even when we run the same experiment in duplicate or triplicate.
Compartamentalized Modules
This simulator was originally created as a tool for developing novel machine learning techniques as applied to upstream process development. It is important not to be able to “cheat” and peer into data to make inferences that are not possible in real life. As such, each module is only able to view data from modules they are connected to. Assays can read environmental conditions and produce a signal that is passed to control strategies, which transforms it into an actuation setpoint, and so on. This aims to ensure that any control or technique employed would work just as effectively in real life as it does here.
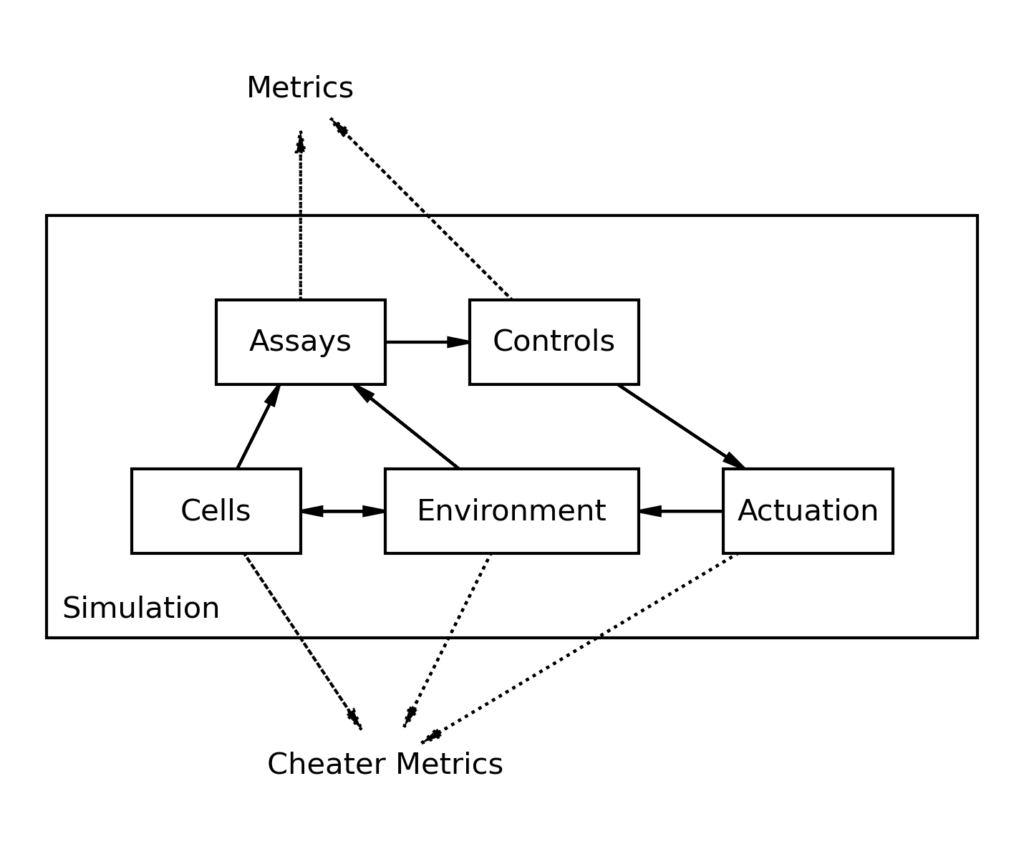
“Cheater” Metrics
While the simulation compartamentalizes data to ensure that data is used appropriately, one of the strengths of simulations is that the exact conditions of the system are known, and can be inspected. If a puzzling situation arises, exact conditions can be inspected to help make sense of it in a way that would not be possible in real life. Concentration of any tracked variable can be graphed with an arbitrary resolution. This could, for example, be used to understand that plummeting pH is due to increased production of lactic acid as a result of a lack of oxygen.
Randomly-Generated Cell Lines
Each cell line is unique, and poses new challenges or optimal parameters to any process. The included CHO-cell model comes with a cell line generator that aims to model this, with each cell line containing over 30 unique parameters that define how the cell line will respond to any given situation. Parameter influence includes:
- Response to varying osmo levels
- Respiratory Quotient
- Shear Sensitivity
- Growth Profile
- Optimal conditions (pH, O2, temperature, etc.)
- PQ profile
- Productivity
Parameters cover a wide range of responses, and can be added and customized for a given variation. This allows for testing how a platform would respond to cell lines that are more or less osmo tolerant, or do not respond to growth inhibition, or are especially shear sensitive.
Units
When programming, it can be easy to overlook something like multiplying by volume, or specifingy a concentration in molar instead of g/L. Especially problematic could be specifying a seed density in e5c/ml instead of e6c/ml. CC-Sim solves this by specifying all parameters passed to class initialization with their units as well as ensuring unit consistency within the simulation. Attempting to add a concentration to a mass will throw an error rather than return erroneous results.
Once the code has been tested with this safeguard, skip_units can be set to True to allow for faster simulations (~50x).
Included Macroscopic CHO-Cell Model
CC-Sim has a specific module for the cell module, and could be adapted to use any host cell, provided a model is supplied. CC-Sim comes with a macroscopic, deterministic CHO-cell model so that the tool can easily used to get a feel for it or cell culture in general.
Care must be taken to understand that the cell model included is a macroscopic rather than a kinetic model of the cell. In a macroscopic model the cell is treated as a black box, and only how the cell responds to conditions are important. As such, the model will not make novel predictions on cell functioning or behave in a way that wasn’t programmed into it. The cell response will only be as good as the equations used to describe it.
This has multiple implications:
- The strength of CC-Sim lies in its modeling of the process, rather than the cells. One could, for instance, set the glucose setpoint of the feed too low; if the process allows for cells to grow unexpectedly or otherwise consume more glucose than is fed, the glucose will run out and this will be reflected in viability. One would not expect, however, to discover that cells will break down HCP for energy reserves; such a thing would need to have a basis in the cell model.
- If a correlation is predicted for cells, it can be input into the model to show how the data would reflect that. For instance, one could compare IGG profiles for cells that have constant volumetric productivity vs cells that have constant cell-specific productivity in order to compare with real data. This would allow for testing of correlations and whether they accurately model a cell.
The cell model is self-contained, and a different cell model can be programmed in or swapped out. Future updates of CC-Sim will likely include more complex and accurate modeling of cells.