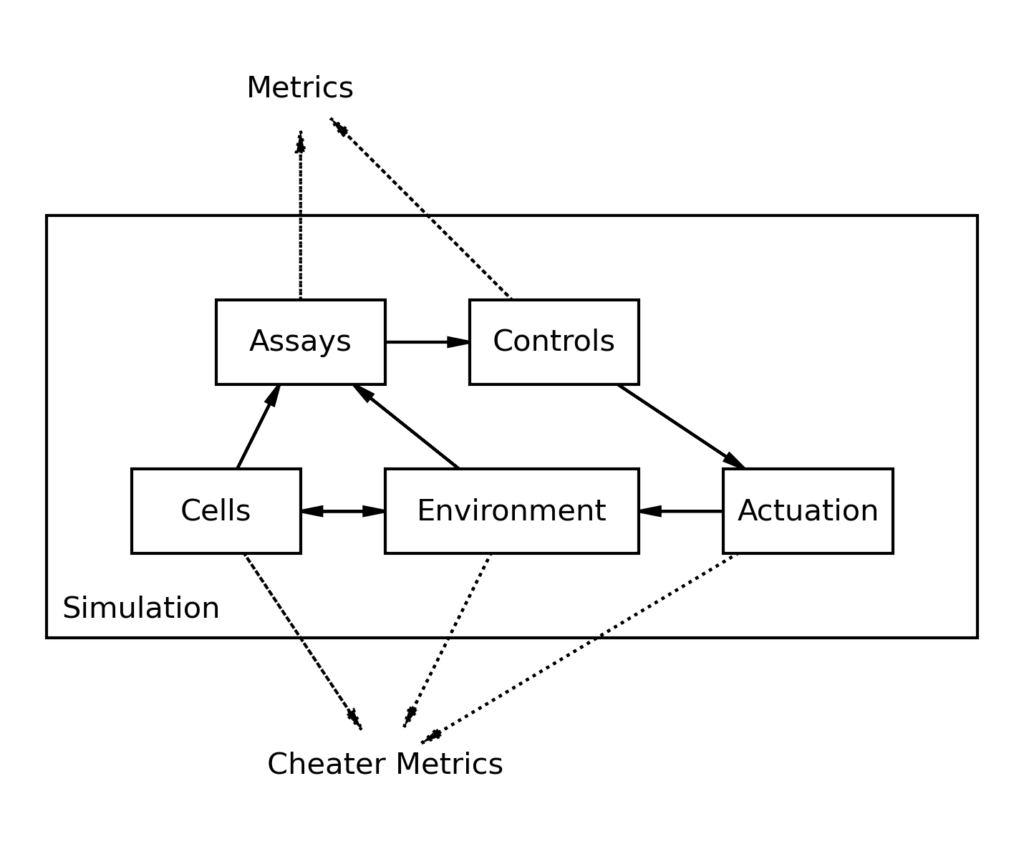
Detailed Design
During the simulation, each step consists of running through each of the below five modules and calling the step() method of their wrapper. The wrapper calculates everything that will happen during the step, and outputs the necessary parameters to the next module so that it may run for a step. You can read about each of the five modules more in detail below:
CC-Sim is a hybrid numerical-analytical simulation. For an in-depth description of what that means and how this is useful, read this workshop post. However, in most cases, it can be assumed that CC-Sim behaves as a continuous simulation where a step size is defined (default: one minute) and every step the simulator cycles through 5 different modules (see above).
Module 1: Assays
Wrapper input: environment, cells
Output: dict of observables for plotting and passing to controls“Assays” are the monitoring equipment that we use to observe the process. It includes everything from online probes (pH, DO) to offline assays like a bioHT or an osmolarity machine. The key functionality of assays is that it has access to the raw state of the simulation (the exact number of cells, or the exact molarity of a component) and then simulates how the assay would act in real life.
For example, the pH probe will start with a systematic error and introduce a random error with each measurement. The probe has both a lag (t98) with its measurement and additionally will drift as time passes.
Each assay class must include:
- Method
read_value(environment, cells), which takes the environment and cell dictionaries as inputs. It must output a dictionary of what it is measuring and the value. - Optionally,
one_point, which allows probes to be one-pointed - The assay configuration is passed to the wrapper, which holds all of the assays configured for an experiment. The wrapper defines whether an assay is online or offline, which dictates whether the assay is run every step or only during an offline sample.
Module 2: Controls
Controls input: assays
Output: dict of metrics (of that step) for later plottingControls have access to every output from assays, and may be programmed to respond in an arbitrary way. The step method of a configured control is called every step, and a dictionary of all observed quantities (from assays) is passed along with a bool offline, which describes whether an offline assay was run that step. Controls may return any metrics (as dict) that they wish the user to be able to graph.
Included in controls is the PID class, which allows for easy configuration of a PID that can be called with step(measured value) and will return the PID value between 0 and 100.
All controls must initialize a list of actuation devices (at self.actuation) that it uses to influence the system. The actuation devices are described in the next module, and contain things such as peristaltic pumps and MFCs.
Controls influence these actuation devices by directly changing their setpoint during each step. For example:
self.actuation[0].set_point = PID_out * self.max_addition_rateModule 3: Actuation
Actuation input: None
Output: A dict of every tracked component describing mass transfer (in mol/s)
in addition to 'RPS', 'heat', 'liquid_volumetric_flowrate', and 'gas_volumetric_flowrate'All devices defined in actuation transform a control signal (the setpoint) into a physical quantity that influences the system. For example, a peristaltic pump could be defined that adds a glucose solution at a rate set by its parent control. Every step, the step() method is called in which the actuation devices calculates and returns the quantity it is influencing the environment with. Each actuation component uses its internal self.set_point (set directly by controls) in order to calculate what occurs during the step.
The quantities should be a rate (e.g. ‘power’ instead of ‘energy’) and treat gaseous and liquids differently.
Gases are specified as a volumetric flow rate. Currently tracked is ‘air’, ‘O2’, and ‘CO2’. They are all assumed to go through the sparger configured in the bioreactor.
Liquids are specified as the total liquid volumetric flow rate ('liquid_volumetric_rate') as well as the molar rate of addition for that component. For example, adding a glucose solution (at 500g/L) could be added as:
{'liquid_volumetric_rate':Q(10., 'ml/min'),
'glucose':Q(27.78, 'mmol/min')}Module 4: Environment
Environment input: actuation, cells
Output: molar concentration of each component in the liquid phase
temperature (in Celsius)
mean shear, max shear
osmolarity
working volume
the time (numpy.datetime64)
pH
maximum consumption rates (if consumption = max, final concentration would equal 0)The ‘environment’ is the environment that the cells are interacting with. The bioreactor serves as its own wrapper (for the simulation) and every call to the step() method the environment calculates and returns the output.
To generate this, the entire bioreactor system (minus actuation) is simulated with inputs from both actuation and the cells.
The environment takes as inputs mass transfer from the cells as well as the actuation, both of which are treated as constants. Mass transfer between the liquid and gas is computed (from solving mass balance differential equations) and the final molarity is returned.
The environment also calculates the maximum average consumption rate possible over a step and passes that to the cell model. If the cell model consumes more than the max consumption rate, it would cause a negative concentration.
The pH calculation assumes that there is bicarb buffer, and that all other acids and bases are strong acids or bases.
Module 5: Cells
Cells input: environment
Output: mass_transfer, a dict of mass transfer rates for each liquid component
(into the cells is positive)
viability
number of living cells (as seen by trypan blue assay)
total cells
diameter
volume (per cell)The cells also serve as their own wrapper for the simulation, and are the most complicated and least understood part of the system, so the exact workings will change. Different models can be swapped out, but must all take the same inputs and return the required outputs. The cells will always take their environment as an input to step() and output quantities needed for the simulation as well as cheater_metrics that allow peering into cells in the simulation in a way not possible in real life.
As of this writing, the included cell model use sigmoidal functions to calculate whether an asymmetric parameter is in range (like shear) and exponential functions for symmetric (osmo). Whether a parameter is in range or not will affect many things, such as extinction rate (cell death), growth rate, target diameter (whether the cells are dividing or just growing in size), and product quality distribution.
Cell models may choose to include internal concentration of components; however, the mass transfer into the cell must never be greater than what is given by max_consumption from the environment; doing so would cause a negative concentration.
Units
When defining quantities, such as the media definition, sparger pore size, or maximum air flow permitted by an MFC, units must be specified. CC-Sim uses the quantities package to handle units. Simply specify a unit by using Q(value, units). A quantity can be converted into SI units by using the method .simplified and rescaled with the .rescale('new units') method.
Example:
from quantities import Quantity as Q
kilogram = Q(1, 'kg')
# A quantity of 1 kilogram is created
kilogram.simplified
# A quantity of 1000 grams
kilogram.rescale('mg')
# A quantity of 1,000,000 milligrams
(kilogram / Q(1, 'min')).simplified
# 16.67 grams per second
kilogram + Q(2, 'mol')
# Error: can't add a mass and molesWhen any class is initialized, it should expected all parameters to be given with units (regardless of whether skip_units is true). Best practice is then immediately converting those units to SI units so that unit conversion only has to be done once.
If skip_units is set to True, all parameters passed between modules (and, preferably, calculations within modules) are done with floats and must be in SI units. This disables the safety and unit checking, but significantly speeds up the simulation (~50x).
float(kilogram)
# Returns 1; wrong
float(kilogram.simplified)
# Returns 1000; correctWhenever changes to the code are made, it is suggested that skip_units be set to False so that an error can be thrown if any mistakes were made. Once a simulation has been run, skip_units can be set to True and the results of each simulation checked against each other. Mistakes can still be made, as functions like math.exp(units) will convert the units to a float in order to perform the exponential; if the units are stored as 200 mmol instead of 0.2 mol, different answers will be obtained. Therefor, care must be taken when using units in an exponent, in a log, or any other mathematical function that is not add, subtract, multiply, or divide.